
The scientific method is based on scientific experiments being verifiable through reproduction. Today, however, some in the scientific community are concerned about the replicability of scientific research. Part of the problem is that given page limits imposed by most publishers, it is generally difficult, if not impossible, to describe scientific methods in sufficient detail to enable the research to be replicated. Furthermore, scientific research is today particularly likely to be viewed as a subject of political debate because there are so many topics of intense concern and interest to the public—from global climate change to genetically enhanced food sources. Those of us who are scientific researchers owe it to each other to make our research as easily replicable as possible. And we owe it to the taxpayers, who fund our research, to minimize the ability of people who critique research to distort its meaning or question the results of research on the basis of politically or financially motivated concerns.
Recently, interesting work has been published related to the replicability of experimental findings, such as an attempt to replicate studies in experimental economic research.1 In this article, I follow earlier distinctions between replicability and reproducibility. By replicability, I mean the ability to replicate an experiment as closely as possible and get generally the same result. The words "as closely as possible" are important: an experiment about an animal population in the wild in a particular area cannot be exactly replicated if the range of the species has moved and is no longer present in the area where an experiment was conducted. On the other hand, if one has the data from a research project, then it should be possible to precisely reproduce the analyses of the data in every detail, extend the analysis if appropriate, and/or correct errors if they exist.
In theory, one should be able to exactly reproduce the data analysis done as part of any research publication. In practice, doing so is hard. Part of the reason this is difficult is that so much of the data analysis is simply not included in published papers, as depicted in figure 1 from James Taylor, one of the creators of the Galaxy bioinformatics software environment.
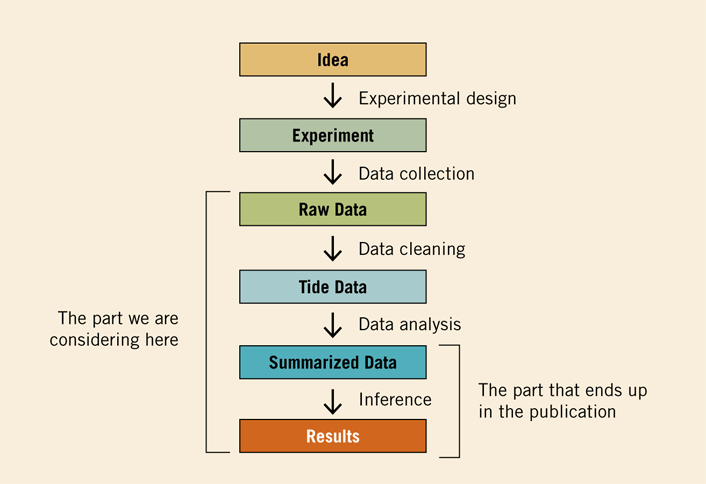
Figure 1. Depiction of Data Analysis Pipeline
Source: James Taylor, "Analysis Reproducibility," Speaker Deck, June 4, 2015 (CC-BY), adapted from J. T. Leek and R. D. Peng, "P Values Are Just the Tip of the Iceberg," Nature 520, no. 7549 (2015), 612.
Another reason it is hard to reproduce scientific data analyses and simulations is because of all the things one must specify in order to make reproducibility possible. Even if you use open-source software, "I used version x of the commonly used open-source software package X" is often all the space one gets. Important details such as which patches, from where, running in what operating system, compiled with what compilers, and using which mathematical libraries are usually omitted. Describing the software environment used for a particular analysis gets very tricky very quickly.
There are very important and useful open-source software repositories available today, notably SourceForge and GitHub. Researchers can make a particular simulation or data-analysis task reproducible by depositing sufficient directions, data sources, copies of output, etc., within one of these repositories. For example, my former (now deceased) colleague Richard Repasky made his published research analyses reproducible by making a .tar file of everything needed to replicate his computer analysis. These .tar archives included the Linux distribution, source code, and make files he used to compile the open-source R software he used for his analysis, along with input data, scripts, and output data. All of these analyses were done with one CPU core in an x86 processor. As long as one can find either an x86 processor or an x86 emulator, one can rebuild the environment he used for any of his published papers and can reproduce his analyses. But doing this takes a fair bit of knowledge and time.
Workflow management systems and virtualization environments are helping to make data analyses and simulations reproducible. Examples of such efforts include RunMyCode, Globus data publication, the Galaxy bioinformatics workflow engine, the Jetstream cloud computing system, and Project Aristotle.
RunMyCode is a highly useful service that enables a researcher to create a companion website to accompany a publication. It is perhaps the most generally available and widely used tool to publish the information, software, and data needed to reproduce scientific data analyses and simulations. However, RunMyCode does not actually provide a place where one researcher can run another researcher's code.
Similarly, the Globus data publication tool makes it possible to publish research data on your own storage systems with standard metadata and curation workflows.
The Galaxy bioinformatics workflow engine is designed to make bioinformatics analyses reproducible, and this initiative has already contributed greatly to the reproducibility of biological research. The website includes a gateway to run a workflow in Galaxy; a workflow run in Galaxy and made available publicly with the relevant data and copies of output should be replicable in other Galaxy workflow portals.
Cloud technology is providing additional new tools for enabling reproducibility of analyses. Jetstream is an example of a science and engineering cloud implemented with enabling reproducibility of scientific research as one of its critical functions. Jetstream is the first production cloud supporting scientific research funded by the National Science Foundation. It uses the Atmosphere interface and is available to any researcher doing open (nonclassified) research at an institution of higher education in the United States. The team operating Jetstream makes available to all users a library of "featured" virtual machines (VMs). These are VMs that provide commonly needed functionality. Jetstream guarantees that they will work properly and, if not, that they will be fixed promptly. There is also a category of VMs that are contributed by one research team for use by others—these are called "contributed" VMs. An individual researcher can use any of these VMs and then store versions—with, for example, data and scripts—in a private VM library. Jetstream thus provides a means by which researchers can distribute scientific software so that other researchers may discover and use it without the headaches of downloading it to a local system.
In addition to the general ability to disseminate and use software within an OpenStack VM, Jetstream provides data-storage and data-publication capabilities that will aid reproducibility of scientific research. Researchers who do analyses with Jetstream may upload a VM image to Indiana University's digital repository (IUScholarWorks) and associate a Digital Object Identifier (DOI) with that VM. This means that researchers have the opportunity to make available a VM containing the entire lifecycle of their research—input data, scripts, all of the programs used to analyze the data, and output data—in a way that allows other researchers to discover that VM through a DOI cited in a paper. A researcher can publish a paper, perhaps with a companion page made via RunMyCode, and provide a VM that any other researcher can download and then run on Jetstream or another OpenStack-based cloud. This provides a very straightforward mechanism for reproducibility of scientific analyses. (Indiana University has committed to make these VMs available "for the foreseeable future"—concretely at least five years past the end of federal funding for Jetstream.)
Project Aristotle aims to take this sort of reproducibility and extend it a step further—making it possible to run VMs across multiple different cloud environments. Already researchers working with Project Aristotle have taken one VM and run it on Cornell University's Red Cloud system, Jetstream, Amazon Web Services, and Microsoft Azure cloud services.
The portability and reusability of VMs provides a practical, straightforward way for researchers to exchange an entire scientific analysis within the confines of a single VM image. This means that scientific software can be preserved as it is used in a particular research project, thus enabling reproducibility of scientific research. In particular, reusability in the form of VM images—with software, data, and results—enables preservation and reusability of software, and thus reproducibility of analyses, in ways that seem relatively manageable to a typical practicing researcher and that will make publication and reuse of data and software a more common and straightforward part of the scientific method as practiced today.
Notes
Jetstream is supported by NSF grant award 144506 and by the Indiana University Pervasive Technology Institute. Any opinions expressed here are those of the author and do not necessarily represent the views of the organizations that have supported research that contributed to this column.
- John Bohannon, "About 40% of Economics Experiments Fail Replication Survey," Science, March 3, 2016.
Craig Stewart is Executive Director of the Indiana University Pervasive Technology Institute and Associate Dean for Research Technologies. He is the principal investigator of the Jetstream project.
© 2016 Craig Stewart. The text of this article is licensed under Creative Commons Attribution 4.0.
EDUCAUSE Review 51, no. 6 (November/December 2016)
